Neuroscientist Spencer Smith, PhD, and his colleagues at the University of North Carolina—Chapel Hill, with the support of the National Science Foundation, have used Zemax OpticStudio™ software to develop new optical systems for two-photon microscopy.
The laboratory of Dr. Smith, including postdoctoral fellow Dr. Jeffrey Stirman, and collaborator Dr. Michael Kudenov of North Carolina State University, worked together to engineer highly customized optical systems that minimize aberrations across large fields of view.
Their work on this project began in September 2014. This microscope has now undergone several iterations, and allows researchers to image brain activity at the single-neuron level—over a much larger area than has ever been possible. Prior to the development of this microscope, individual neurons had been viewed only in very small portions of single brain regions.
The new microscope enables extremely high-resolution two-photon imaging of multiple brain areas, retaining the resolution that allows observation of individual neurons. The information this provides is new, precious, and ripe with untapped potential.
Dr. Smith and his colleagues used Zemax OpticStudio™ to model optical systems and optimize performance to achieve the stunning, unprecedented ability to maintain and even increase resolution over these much larger areas.
Thanks to the software’s extensive optimization tools and stock lens matching tools, these scientists were able to rapidly develop custom optical systems for a next-generation two-photon microscope that is literally changing how we see the brain.
This research was funded in part by the White House’s BRAIN (Brain Research through Advancing Innovative Neurotechnologies) Initiative—a bold new research effort to revolutionize our understanding of the human mind and uncover new ways to treat, prevent, and cure brain disorders (such as Alzheimer’s, schizophrenia, autism, epilepsy, and traumatic brain injury).
The multi-institute initiative was also supported by both federal and private foundations. The National Science Foundation and Zemax have supported the advanced optical instrument development by Dr. Smith, and the Simons Foundation is supporting the groundbreaking neuroscience research that takes advantage of the new microscope. A McKnight Foundation award is also funding a new imaging instrumentation project that has grown out of this earlier work.
We talked to Dr. Smith last month, one year after the announcement of the project and its funding, to explore the passion that prompted the project; how the process unfolded; the role OpticStudio™ played; the excitement of being able to image individual neurons over multiple brain areas; and the radical possibilities that extend from this breakthrough.
Zemax: What led you to develop this particular microscope, and why was it important to you?
Dr. Smith: I’m interested in how neurons, and the networks they form, compute. As we explore that in our research, some experiments are just on individual neurons. But in others we want to look at an entire population of neurons and measure their activity. We then want to correlate this activity with the types of stimuli that the animals are experiencing, and the animals’ subsequent behavior. This way, we can start to understand how stimuli and behavior are encoded in the intricate patterns of neuronal activity distributed across many brain areas acting in concert.
Neural activity in the visual cortex represents everything we see. In experiments with mice, we show the mice different types of videos and measure neural activity in the brain in response to that stimuli. We examine how the brain encodes that stimuli, and how it sets up its circuitry in development to do so.
Historically, we’ve been able to image all this only in one little portion of the brain at a time.
During my postdoctoral work in London, I was doing an experiment and reflecting about how one of the amazing features of the brain is that it has all these different areas that work together in complex ways. The primary visual cortex sends information to other areas: higher visual areas that encode special features of visual stimuli.
So there’s initial, processing at one level that we’re observing, but simultaneously there is additional processing at other levels that we don’t yet understand. And it’s difficult to determine how these levels work we don’t look at all the activity, in all of the areas, at the same time.
One of the best instruments we had was two-photon microscopy. While this approach provides single-neuron resolution, it was only letting us see one little portion of the brain at a time. I wanted to get a larger field of view. The classic way to do that is to use a microscope objective with smaller magnification.
Unfortunately, because microscope objectives tend to be small, a smaller magnification would necessarily also involve lower resolution. Thus although this standard solution can indeed allow one to image multiple brain areas, the resolution is so low that individual neurons are blurred together.
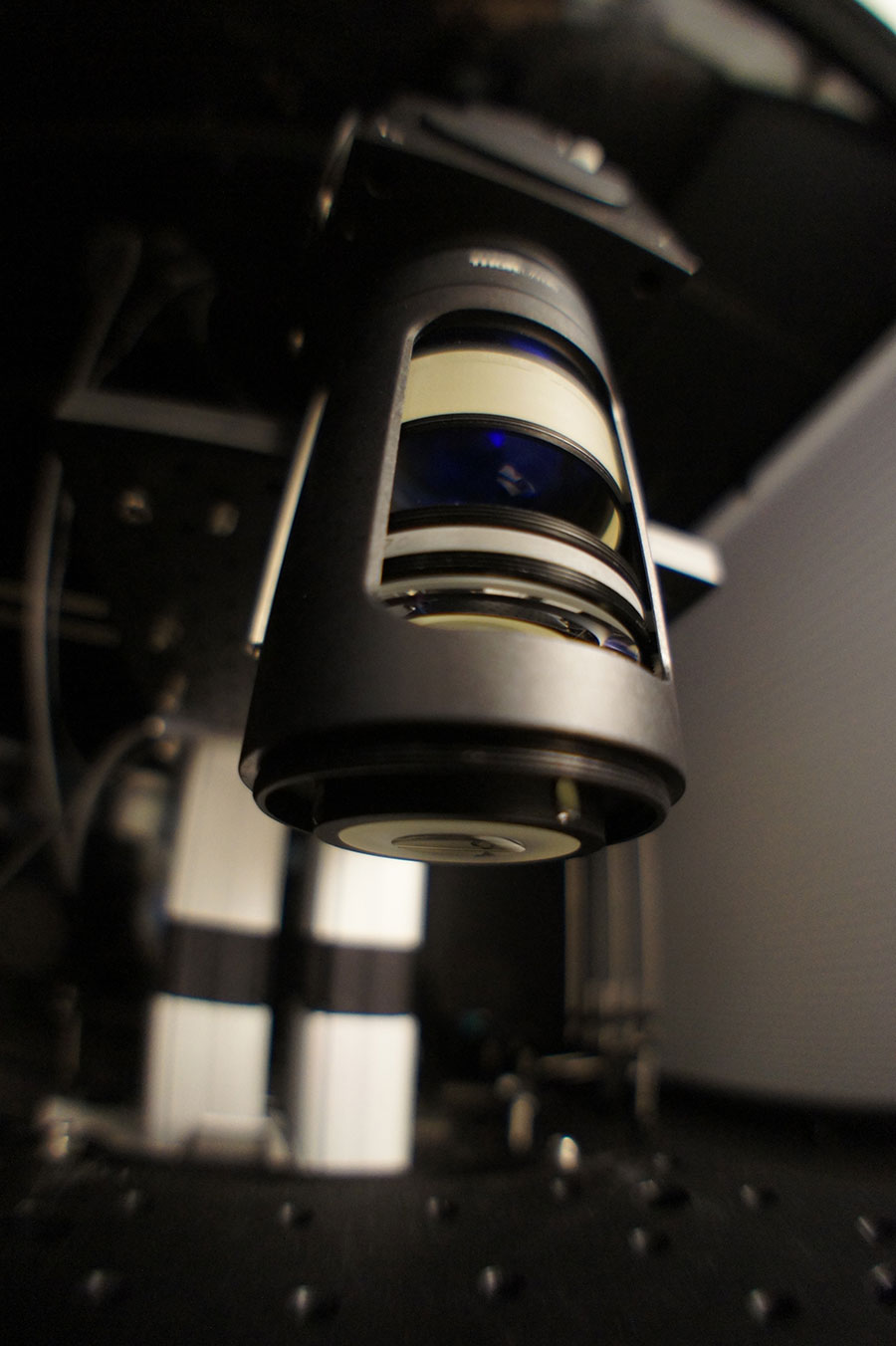
Obviously, we didn’t want to make that compromise. We wanted to distinguish individual neurons, but over a larger field of view—because that would give us exciting new information that could illuminate how brain areas work together. The coordination of activity in multiple brain areas is essential for brain function, and this coordination may be involved in the pathology of some complex neurological diseases.
That’s the essential problem that drove this development—wanting to measure brain activity in multiple areas at the same time, in the same field of view, and monitor individual neurons. And the instrument simply didn’t exist.
Zemax: So how did you approach the development of a microscope with capabilities that didn’t yet exist?
Dr. Smith: Well, I thought about how the optical components fit together. Simply designing a new microscope objective would not be sufficient. Two-photon microscopy uses a scanning laser to image cells. So we had to have a scan engine that could scan a large-diameter laser beam at relatively high angles. Optical scan engines for a large field of view have to be specially engineered. It was a really big project.
I won critical early support in the form of a Career Development Award from the Human Frontier Science Program (HFSP). It was transformative for my research program to have this risky project funded early in my career. With this new funding, I contacted companies that do this kind optical design and fabrication.
I gave those companies some initial specs, and it quickly became clear that it would take six months to a year, maybe longer, and tens of thousands—if not hundreds of thousands!—of dollars to get one version of this system developed. And if I didn’t get all the parameters just right the first time, I’d be pulling the trigger on a lot of money for something that might not work. I wouldn’t have the time and resources to do another iteration.
In the background, to complicate matters further, lots of changes were taking place concurrently in laser technology, and in the different kinds of genetic engineering we use to label neurons. We use genetically engineered proteins to optically label neurons and report neural activity. Those engineered proteins were undergoing rapid development as well. To decide a priori what optical parameters we needed, and give those to a lab, with so many factors in flux…it was highly unlikely that in one shot, we’d end up with what we needed.
What all that meant was that, in order to have the latitude and flexibility to experiment, and without spending prohibitive amounts of time and money, I’d need to do this development in-house. By doing it ourselves, with the right software, we could iterate extensively to see which engineering compromises we could make.
So, thanks to our grant funding and Zemax and other support, we began designing and building the system ourselves.
It was amazing. This approach worked even better than I expected—way better. I was hoping for a two-fold, maybe three-fold increase in the field of view. We got a 50-fold to 100-fold increase in the imaging area. It was breathtaking. Like taking blinders off for the first time.
Zemax: We gather that this is where OpticStudio™ came in—allowing you to effectively create this microscope yourself rather than “farm it out.”
Dr. Smith: Yes. There were several ways that Zemax software was essential to our in-house development of this new instrument. First, OpticStudio™ has a stock lens matching tool. That was a tremendously helpful tool to work with. We could set up the performance we wanted, and the software tries a bunch of variations to get you what you want. We can ask it to design any lens under the sun, or stick to the catalog. We could get an optical system design done, order the stock lenses into the lab within 24-48 hours, try it out, and determine additional parameters we needed to hit. Even with custom lenses, we could restrict the optimization to use materials that we knew our favorite lens maker has in stock. And with the quoting tool, we could find out if something was going to cost $10,000 vs. $100,000 to actually build, which of course is critical.
OpticStudio™ also has a really nice merit function editor—definitely a strength of the software. When you set out to sketch an optical layout and have the software optimize it, this function assigns a score to the solution. The score can be related to the optical transfer function, the RMS wavefront error, or other properties. Also, the design can be micromanaged to an extremely fine degree. This was fantastic, and this is really where the magic happens in optical design. The quality of the solution that the software finds can be surprisingly sensitive to differences in the merit function, so it’s great to have fine and easy control.
In addition to the support of Zemax and the software, this project was a team effort. Most of the work was done by Jeffrey Stirman, Ph.D., a post-doc in my lab. He’s a bioengineering PhD, and recently won a prestigious Burroughs Wellcome Career Award at the Scientific Interface. He is our “merit function maestro” and as such, really helped us squeeze performance out of everything we designed and get fast turnaround.
Another key player is Michael Kudenov, Ph.D., an assistant professor at North Carolina State University. He’s the optics expert who was instrumental to getting us started the right way. He’s the one who got us using Zemax software in the first place; he designed an early version of the custom objective—really helping us frame and think about the problem correctly and develop the early prototypes—and he continues to give us valuable guidance in this work.
Zemax: Tell us what happened when you actually had a working instrument that met your parameters.
Dr. Smith: Well, it was amazing. This approach worked even better than I expected—way better. I was hoping for a two-fold, maybe three-fold increase in the field of view. We got a 50-fold to 100-fold increase in the imaging area. It was breathtaking. Like taking blinders off for the first time.
We could zoom out and see individual neurons—key to our experiments—and see it over a 9.6 mm2 area. In mice, that’s pretty much the entire primary visual cortex plus six or seven higher visual areas. This opens up whole new unexplored territories in neuroscience experiments.
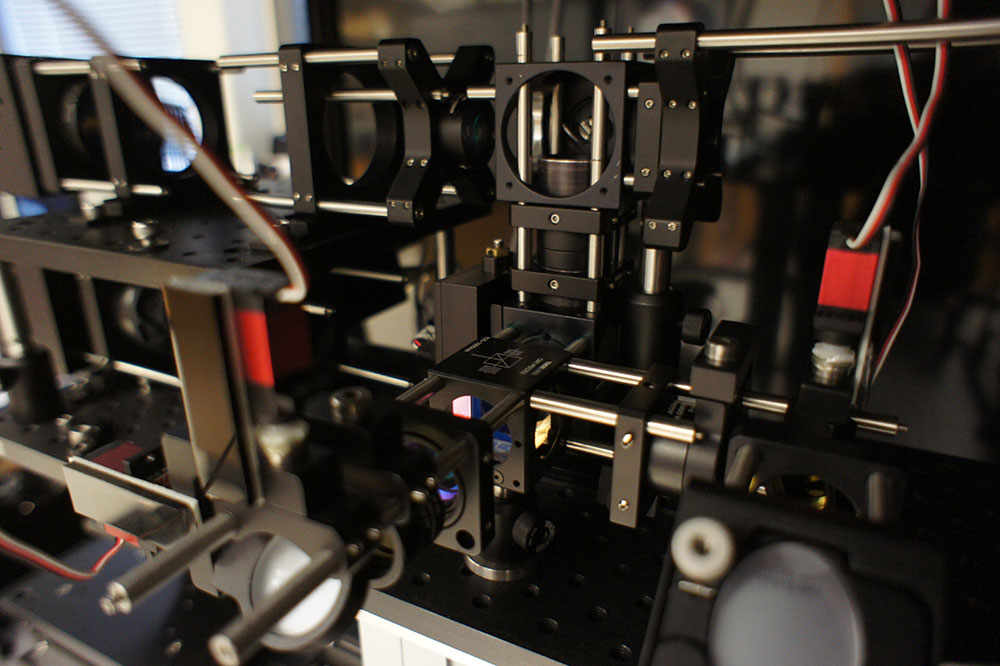
Zemax: Tell us a little more about the kinds of experiments you’re doing now that you have this unsurpassed tool. What might these experiments ultimately offer for humans?
Dr. Smith: One way to think about how the brain processes stimuli is to think about a telephone conversation. We have parts of the brain talking to each other, all processing input—including, for example, visual input. These parts have interconnections and exchange information.
If you only look at or hear one side of a phone call, you don’t know what other side is doing with the information. It can be difficult to decipher meaning or context or draw conclusions, unless you listen to both sides.
When we listen to activity in cortical areas we know are connected, we can perform statistical analysis based on which areas respond when. With one visual stimulus, it appears that brain areas A and B talk and exchange a lot, but A and C do not. With a different stimulus, A and C talk extensively, but then A and B do not.
These are the kinds of questions we can now ask. Our first experiments involved making measurements, and it turned out to be a lot more dynamic than we initially expected. We can work with behaving animals and see how different parts of the brain work in concert depending on demands, learning, and performance.
It may sound abstract, but in practical terms, this gives us a better understanding of how different parts of brain work together to drive behavior. And that aspect of brain function is likely a piece of the puzzle in complex neurological diseases such as schizophrenia and autism—ones that don’t necessarily involve “sick cells” or neurodegeneration, but rather a circuitry issue or dysfunctional plasticity.
Observing this kind of “network activity” over its entirety enables clinical researchers to identify a locus of dysfunction. It can show us what regulates processes normally, and what may may fail to develop normally in cases of disease.
So much of how we’ve understood these diseases historically has been focused on individual synaptic connections and the molecules involved in them. But to come up with a new macroscopic-level understanding is vital. Conventional models are great at telling us what’s dysfunctional in a cell or synapse, but at a network level...long-range circuitry and dynamics...we really don’t know much. Now we have a new tool to investigate this level of brain circuitry.
For human diseases linked to a single gene, we can mutate that gene in a mouse model and ask what happens to mammalian circuitry when that gene is disrupted. We can get an idea of what mechanisms are important to ensure the proper development and function of that circuitry, and what happens to it during learning and behavior. We can understand how human diseases both are caused by, and can cause disruptions in, brain circuitry.
Ultimately what these experiments might mean for people is new therapeutic strategies—discoveries of druggable targets that weren’t considered earlier, or looked at in the right way. It also may give us ideas to ensure proper development post-natally.
We’re sharing all of this data, of course, with the BRAIN Initiative.
I’m also very interested in computation. One of the things a lab mouse does better than any system we design is process visual input. They do it faster and more efficiently than any of the latest computer vision algorithms. So I’m curious about the computation that’s designed into that circuitry—what it is that enables such efficiency and speed.
We don’t yet understand everything about how that works, what affords a brain its speed. And again, we think one key component is multiple brain areas working in concert—which we can now view. It’s underexplored, and when we fill in the gaps of understanding about how the brain processes visual information, we’ll be able to develop better algorithms for better artificial intelligence. So that’s highly speculative, but if I’m dreaming large…that’s a huge area of potential.
We could zoom out and see individual neurons—key to our experiments—and see it over...pretty much the entire primary visual cortex plus six or seven higher visual areas. This opens up whole new unexplored territories in neuroscience experiments.
Zemax: Can this microscope be made even better?
Dr. Smith: We’re working on that. What we have right now is already beyond what I set out to build with the first grant I received, from HFSP. The first time around, we used stock lenses, but more recently, with NSF support, we have revised the optics with custom lenses to squeeze all the performance we could out of the system.
Now we’re pushing the envelope, trying to get an even larger field of view and higher resolution, and optimize various parameters for specific experiments. We’ve currently “frozen” the first system, so it is now dedicated to generating data. We have expanded our lab space and we’re putting a new imaging system together that will be even better.
I thought this was going to be a one-shot deal. But then the BRAIN Initiative came out a year or two into this project, after we were already working with money from the HFSP grant. The initiative identified as a key area of focus the development of better tools and technology—better tools for measuring brain function in model systems.
There’s usually not much money in biology to develop new tools. The NIH funds most biological work in the US, and it tends to fund experiments rather than tool development. So this was a great new opportunity.
Now, we’ll see how far we can push this technology in our own lab.
We’re also trying to move forward in a way that makes it easy for other labs to adopt what we’re doing. We want other neuroscientists to replicate and expand on our experiments. Not everyone wants the same optical parameters, so ultimately we hope different variations will evolve from this platform that can satisfy the needs of many different experiments. We’ve explored this with some smaller microscope manufacturers.